Research Report
Screening and validation of Hepatocellular Carcinoma Related Genes Based on GEO Database 
 Author
Author  Correspondence author
Correspondence author
International Journal of Molecular Medical Science, 2020, Vol. 10, No. 4
Received: 14 Feb., 2020 Accepted: 27 Mar., 2020 Published: 27 Apr., 2020
Hepatocellular carcinoma (HCC) is one of the most common malignant tumors in China. Identifying the genes related to the development of HCC is of great significance for the in-depth study of the pathogenesis of HCC and the development of therapeutic targets. The GEO2R tool was used to screen the differentially expressed genes common to five data sets from the Gene Expression Omnibus Database (GEO). Those differentially expressed genes were potential hepatocellular cancer-related genes. Functional enrichment and signal pathway analysis of differentially expressed genes were carried out by Metascape. The Gene Expression Profiling Interaction Analysis (GEPIA) was used to identify gene closely related to clinical significance of HCC patients. Real-time fluorescence quantitative PCR was used to verify the differentially expressed genes related to the prognosis of liver cancer and to identify the candidate genes related to liver cancer, laying a solid foundation for further research. It was found that A total of 94 differentially expressed genes were shared by the 5 data sets. After literature search, it was found that the relationship between 24 genes and the development of HCC was rarely reported in literature, and they were unknown functional genes in HCC. After analyzing the data in The Cancer Genome Atlas (TCGA) with the GEPIA. It was found that GINS1 was highly expressed in liver Cancer tissues and negatively correlated with survival of liver cancer patients. CFHR4 and DNASE1L3 were significantly lower expressed in liver cancer tissues and were positively correlated with survival of liver cancer patients. Real-time fluorescence quantitative PCR confirmed that GINS1 was highly expressed in 81.3% HCC tissues, while CFHR4 and DNASE1L3 were respectively low expressed in 71.9% and 93.8% HCC tissues. Therefore, it was found that GINS1, CFHR4 and DNASE1L3 were significantly differentially expressed in HCC tissues, which are closely associated to the prognosis of HCC patients, and may be used as potential molecular markers to diagnose the prognosis of HCC patients and potential drug targets for the development of HCC treatment.
Hepatocellular carcinoma is one of the most common malignant tumors and the sixth most common cancer in the world, with the death rate ranked fourth among cancers (Bray et al., 2018). HCC ranks 5th among the top 10 high-incidence cancers in China. And it ranks 3rd in lethality in China, after lung cancer and gastric cancer (Chen et al., 2016). At present, although there are various methods for the treatment of HCC, such as hepatectomy, liver transplantation, and molecular targeted drugs (sorafenib), due to the high recurrence and high metastasis of liver cancer, the therapeutic effect is still not ideal (Bruix et al., 2014; Reig et al., 2016; Rossetto et al. 2019). Therefore, the search for genes related to the diagnosis and development of HCC is of great significance for the in-depth study of the pathogenesis of HCC and the development of diagnostic and therapeutic techniques.
The Gene Expression Omnibus Database (GEO) contains more than 32 000 public data sets from 13 000 laboratories, including data from multiple platforms such as gene chips, next-generation sequencing, and high-throughput sequencing. And it provides important support data for multi-sample research of tumors (Barrett et al., 2013). More and more researches are based on data mining conducted on the GEO, and many experiments have confirmed that the key genes discovered have an important role in the occurrence and development of cancers (Shen et al., 2019; Ye et al. 2019). This study uses the GEO data sets and a variety of bioinformatics analysis methods to find molecular markers that are closely related to the occurrence and development of HCC, providing a new basis for the diagnosis and treatment of HCC.
1 Results and Analysis
1.1 Screening of differentially expressed genes
GEO2R is an online analysis tool. After analyze two or more groups of data in the GEO database, it can obtain differentially expressed genes (DEGs). Use the GEO2R to analyze the GSE60502, GSE101685, GSE84402, GSE29721, and GSE33006. After that, selected the genes within the range of | log2FC | and p<0.05. The analysis results show that 5 827 differentially expressed genes were obtained in the 5 datasets. And the top 50 genes with the most obvious differences in each data set can distinguish between cancer and non-cancer samples (Figure 1A). In addition, there are 94 DEGs shared in the 5 datasets, including 28 up-regulated genes and 66 down-regulated genes (Figure 1B).
|
Figure 1 Differential expression of GEO data sets Note: A: Heatmap with top 50 genes in five GEO data sets; B: Venn diagram of differentially expressed genes in five GEO data sets |
1.2 Enrichment analysis for DEGs
Metascape is an online website that integrates a variety of biological analyses such as gene annotation, functional enrichment analysis, and signal pathway analysis (Zhou et al., 2019). Using the Metascape performed functional enrichment and signaling pathway analysis on 28 up-regulated genes and 66 down-regulated genes related to HCC. It was found that the functions of up-regulated genes are mainly enriched in biological processes such as cell division, regulation of cell cycle process and chromosome condensation. And the signaling pathway is mainly related to the cell cycle and p53 signaling pathway (Figure 2A). The functions of down-regulated genes are mainly enriched in the biological processes of acute inflammation response, steroid metabolism process and cellular response to copper ion. And the signaling pathways are mainly related to retinol metabolism, mineral absorption, complement and coagulation cascades (Figure 2B).
|
Figure 2 Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis Note: A: Up-regulated GO and KEGG analysis; B: Down-regulated GO and KEGG analysis |
1.3 Screening of genes related to liver cancer
National Center for Biotechnology Information is currently the largest medical and scientific literature retrieval database in the world. GEPIA is an online interactive tool that analyzes the expression of a gene in cancer, its correlation with patient survival, and other biological information (Tang et al., 2017).
In this study, NCBI was used to conduct a literature search on 94 differentially expressed genes. And it was found that 70 genes have reported related molecular mechanisms in the development of HCC, and 24 genes have not been searched for in-depth reports on the molecular mechanisms related to the development of HCC. Twenty-four genes that were not thoroughly studied were initially identified as candidate genes for subsequent analysis, including 2 up-regulated genes and 22 down-regulated genes (Table 1).
|
Table 1 Ninety-four differentially expressed genes (DEGs) associated with hepatocellular carcinoma (HCC) were screened from GSE60502, GSE101685, GSE84402, GSE29721 and GSE33006 microarrays |
After analyzing the data in TCGA by GEPIA, the expressions of the 24 DEGs that were not thoroughly discussed are consistent with the analysis results of this study (Figure 3A). And GEPIA was used to analyze the association of these genes with the prognosis of HCC patients. It was found that only GINS1, CFHR4, and DNASE1L3 were significantly correlated with the overall survival and disease-free survival of liver cancer patients (Figure 3B).
|
Figure 3 Expression of candidate genes in HCC samples and correlation analysis of some genes with patient survival Note: A: Expression of 24 candidate genes in HCC samples, the red is tumor tissue (Tumor, T, n=369), while the gray is normal tissue (Normal, N, n=50). Statistical significance (*p<0.01) is indicated for cancer tissues compared with the para-cancer tissues; B: The analysis of GINS1, CFHR4 and DNASE1L3 with overall survival and disease-free survival of HCC patients, p(HR)<0.05 indicates that this gene is associated with the prognosis of HCC patients |
1.4 Candidate gene verified by real-time fluorescent quantitative PCR
For the GINS1, CFHR4 and DNASE1L3 screened above, real-time fluorescence quantitative PCR was used to verify the expression level of 32 liver cancer and para-cancer samples. GINS1 was highly expressed in 81.3% of HCC tissue samples, CFHR4 and DNASE1L3 were lowly expressed in 71.9% and 93.8% of HCC tissue samples, respectively. The verification results of GINS1, CFHR4 and DNASE1L3 in HCC tissues are consistent with the analysis results (Figure 4).
|
Figure 4 The expression of GINS1, CFHR4 and Note: Results are shown as the mean±SD (n=32); Statistical significance (***p<0.001 or ****p<0.0001) is indicated for cancer tissues compaDNASE1L3 were detected by real-time quantitative PCR in 32 pairs of HCC and adjacent samplesred with the para-cancer tissues |
2 Discussion
At present, many studies have shown that DEGs in patients with HCC have a great effect on the occurrence and development of HCC (Huang et al., 2007; Xia et al., 2016; Yuan et al., 2019). However, the biomarkers that can be used for clinical diagnosis are mainly AFP. But used AFP alone for diagnosis has a high false negative rate (Yu et al., 2019). Therefore, biomarkers for diagnosis and treatment of HCC are still in the exploration stage. So, we need more researches to explore related genes that have an important role in the development of HCC. In this study, bioinformatics was used to analyze the data of 5 datasets (GSE60502, GSE101685, GSE84402, GSE29721, GSE33006) on the GEO database. And the bioinformatics analysis was carried out to find the unknown functional genes closely related to the development of HCC. These genes may lay the foundation for in-depth study of the pathogenesis of HCC and the development of new diagnostic and therapeutic targets.
By analyzing 5 datasets, a total of 94 common DEGs were obtained (28 up-regulated genes and 66 down-regulated genes). Subsequent functional enrichment analysis found that the functions of up-regulated genes are mainly enriched in biological processes closely related to cancer, such as cell division and cell cycle; down-regulated genes are mainly enriched in many biological metabolic processes, such as acute inflammation response, steroid metabolism process and cellular response to copper ion. At present, many studies have confirmed that mitotic abnormalities play an important role in the development of cancer. And inhibitors of many genes related to mitosis have entered the stage of clinical testing, hoping to play a role in cancer treatment (Dominguez-Brauer et al., 2015). Studies have shown that the occurrence of cancer is accompanied by changes in many metabolic pathways in the body, and these metabolic pathways may play an important role in the occurrence and development of cancer (DeBerardinis et al., 2008). This suggests that the key genes involved in the metabolic pathway and its pathway may be closely related to the occurrence and development of HCC. It may provide new ideas for the diagnosis of HCC. Analysis of the signal pathways of DEGs showed that the DEGs related to HCC in this study were mainly involved in the cell cycle, p53, retinol metabolism, mineral absorption, and complement and coagulation cascade pathways. The p53 signaling pathway is one of the most important apoptosis signaling pathways. Many genes in this pathway, such as TP53, play an important role in the development of HCC (Lai et al., 2007). The retinol metabolism pathway mainly is one of the pathways involved in liver fibrosis. Some studies have shown that the enzymes involved in retinol metabolism are related to HCC. Many genes in the pathway play an important role in occurrence and development, such as CYP1A2, CYP2C9 (Yu et al., 2015; Ren et al., 2016). In addition, some genes in metabolic pathways such as mineral absorption and complement and coagulation cascades have also been shown to be involved in the development of HCC (Seol et al., 2016; Liu et al., 2018). Some scientists have suggested that patients always with the change of multiple metabolic pathways during the development of HCC. Inhibiting key genes of tumor metabolism may become a new method of tumor treatment (Monte et al., 2005; Wu et al., 2010; Luengo et al., 2017). After analyzed by GO and KEGG, we found that the 94 candidate genes were play an important role in the occurrence and development of HCC. The key genes which involved in HCC metabolic pathways in the 94 candidate genes are likely to become targets for the treatment and diagnosis of HCC especially.
This research is to further study the molecular mechanism of the occurrence and development of HCC and find important genes with unknown functions in the pathogenesis of HCC. We conducted a literature search on 94 differential genes and found that 70 genes (such as TOP2A, CDK1, BIRC5, etc.) have been reported in the literature to be closely related to the development of HCC (Wong et al., 2009; Cao et al., 2013; Wu et al., 2018). But there are still 24 genes that have not been found to be related to the molecular mechanism of HCC in-depth research reports. Therefore, we listed 24 genes that were not reported in-depth as candidate genes, and conducted follow-up analysis. Using the GEPIA to verify the expression of 24 genes, it was found that 24 genes all showed differential expression in HCC. And the expression pattern was consistent with the analysis results. This further suggests the reliability of our analysis results. Further analysis found that only 3 of the 24 genes (GINS1, CFHR4, and DNASE1L3) were closely related to the disease-free survival and overall survival of HCC patients (p<0.05). In order to better verify our results, we used real-time fluorescence quantitative PCR technology to detect the expression differences of these three genes in 32 pairs of HCC and HCC adjacent tissue samples. The results showed that GINS1 had high expression in 81.3% of HCC tissues, CFHR4 and DNASE1L3 were down-regulated in 71.9% and 93.8% of HCC tissues, respectively.
The down-regulated of CFHR4 and DNASE1L3 and the up-regulated of GINS1 in HCC tissues found in this study has not been reported. Some scientists have found that CFHR4 may be associated with C-reactive protein (CRP). CRP can be used as an inflammatory marker that reflects the growth and invasion of HCC patients. This suggests that in-depth study of the relationship between CFHR4 and CRP may find new targets for liver cancer treatment (Mihlan et al., 2009; Suner et al., 2018).
In summary, this study found three HCC-related genes and verified their expression differences in HCC tissues. Besides, we initially discussed the relationship between the three HCC-related genes and the prognosis of HCC. Our research results suggest that in-depth study of the role of these three genes in the pathogenesis of HCC may find new molecular targets for diagnosis and treatment of HCC and provide a certain research basis for the development of diagnosis and treatment technology.
3 Materials and Methods
3.1 Experimental reagents
PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time) and TB Green® Premix Ex Taq™ (Tli RNaseH Plus) were purchased from TAKARA. The reagents were used for reverse transcription of sample RNA and determination of the relative expression of genes in the sample.
3.2 Microarray data
GSE60502, GSE101685, GSE84402, GSE29721, and GSE33006 were obtained from the GEO database. GSE60502 is a dataset based on the GPL96 platform submitted by Kao K.J. et al. It includes 36 samples from HCC and normal liver tissue of 18 patients. GSE101685, GSE84402, GSE29721 and GSE33006 are datasets based on the GPL570 platform. Among them, GSE101685 was submitted by Sen-Yung H. et al., including 8 normal samples and 24 HCC samples. GSE84402 was submitted by Qin W. et al., including 28 samples from 14 patients. GSE29721 was submitted by Bhattacharyya B., etc., including 10 normal samples and 10 HCC samples. GSE33006 was submitted by Huang Y. et al., including 3 HCC samples and 3 normal samples.
3.3 Screening of DEGs
The online analysis tool GEO2R was used to analyze DEGs (https://www.ncbi.nlm.nih.gov/geo/geo2r/). The DEGs between HCC tissues and normal HCC tissues were screened DEGs with the threshold of |log Fold Chang(FC)|≥2 and p<0.05. After that, we used Morpheus (https://software.broadinstitute.org/morpheus/) to raw a heat map of the TOP50 DEGs in 5 datasets. In addition, we used the R language to draw the Venn map of the DEGs obtained from the 5 datasets to obtain the DEGs shared by 5 datasets.
3.4 Functional enrichment analysis
The online analysis tool Matescape (http://metascape.org/) performs GO enrichment and KEGG analysis on the DEGs shared by the five datasets. Screening biological functions and signaling pathways with p<0.01 and at least 3 genes involved in.
3.5 Screening of DEGs related to HCC
The DEGs related to HCC were searched with NCBI (https://www.ncbi.nlm.nih.gov/). In order to find the genes that had no in-depth reports on molecular mechanisms related to HCC. Subsequently, the online analysis tool GEPIA (http://gepia.cancer-pku.cn/) was used to analyze the expression levels of DEGs related to HCC, as well as the correlation with the patient's overall survival and disease-free survival. Genes with a correlation between survival and disease-free survival are listed as candidate genes.
3.6 Real-time quantitative PCR verification
The candidate genes screened above were verified by real-time fluorescence quantitative PCR. Take 1 µg sample RNA for reverse transcription and follow the instructions of PrimeScript™ RT reagent Kit with gDNA Eraser (Perfect Real Time) to obtain cDNA. The real-time fluorescence quantitative PCR instrument StepOnePlus is used for quantitative PCR analysis and the subsequent steps and program settings are operated according to the TB Green® Premix Ex Taq™ (Tli RNaseH Plus) instructions. In the experiment, β-ACTIN was used as the internal reference gene, and 2-ΔΔCt was used to calculate the relative expression level.
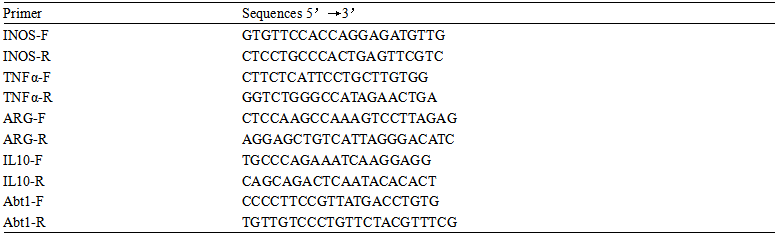
The real-time fluorescence quantitative PCR primers are as follows:
β-ACTIN Forward primer: 5'-CACCAACTGGGACGACAT-3', Reverse primer: 5'-ACAGCCTGGATAGCAA CG-3'; GINS1 Forward primer: 5'-CCCAGATACCATTTTGGCGTG-3', Reverse primer: 5'-GTCTGAGTCCA TCCTCGTTG-3'; CFHR4 Forward primer: 5'-TCAAACCCCAAACAGTGCAAC-3', Reverse primer: 5'-ACA AGGTTTCACTTCTTGTCCA-3'; DNASE1L3 Forward primer: 5'-TCAACGTCAGGTCC TTTGGG-3', Reverse primer: 5'-GCTTTTCCCTGTTCAGCTTCTC-3'.
3.7 Statistical analysis
GraphPad Prism 5.0, Microsoft Excel and Microsoft PowerPoint were used for data and graph processing. The data were expressed in the form of mean ± standard deviation in statistical analysis. The difference between cancer and adjacent samples was calculated by unpaired t test, p<0.05 indicates statistical significance.
Authors’ contributions
Qiu Lin is responsible for the entire data analysis, experimental operation and thesis writing; Professor Huang Jian is the main person in charge of this research, responsible for the design of the experimental ideas, technical guidance and thesis modification. Both authors read and agreed to the final text.
Acknowledgement
This research was jointly funded by the Natural Science Foundation Commission Fund (No. 18Z103010173) and the key project of Shanghai Science and Technology Commission (No. 19Z111220022).
Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., Marshall K.A., Phillippy K.H., Sherman P.M., Holko M., Yefanov A., Lee H., Zhang N., Robertson C.L., Serova N., Davis S., and Soboleva A., 2013, NCBI GEO: archive for functional genomics data sets—update, Nucleic Acids Research, 41(Database issue): D991-D995
https://doi.org/10.1093/nar/gks1193
Bray F., Ferlay J., Soerjomataram I., Siegel R.L., Torre L.A., and Jemal A., 2018, Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries, CA: A Cancer Journal for Clinicians, 68(6): 394-424
https://doi.org/10.3322/caac.21492
PMid:30207593
Bruix J., Gores G.J., and Mazzaferro V., 2014, Hepatocellular carcinoma: clinical frontiers and perspectives, Gut, 63(5): 844-55
https://doi.org/10.1136/gutjnl-2013-306627
Cao L., Li C., Shen S., Yan Y., Ji W., Wang J., Qian H., Jiang X., Li Z., Wu M., Zhang Y., and Su C., 2013, OCT4 increases BIRC5 and CCND1 expression and promotes cancer progression in hepatocellular carcinoma, BMC Cancer, 13: 82
https://doi.org/10.1186/1471-2407-13-82
PMid:23433354 PMCid:PMC3583731
Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F., Jemal A., Yu X.Q., and He J., 2016, Cancer statistics in China, 2015, CA: A Cancer Journal for Clinicians, 66(2): 115-132
https://doi.org/10.3322/caac.21338
PMid:26808342
DeBerardinis R.J., Lum J.J., Hatzivassiliou G., and Thompson C.B., 2008, The Biology of Cancer: Metabolic Reprogramming Fuels Cell Growth and Proliferation, Cell Metabolism, 7(1): 11-20
https://doi.org/10.1016/j.cmet.2007.10.002
PMid:18177721
Dominguez-Brauer C., Thu K.L., Mason J.M., Blaser H., Bray M.R., and Mak T.W., 2015, Targeting Mitosis in Cancer: Emerging Strategies, Molecular Cell, 60(4): 524-536
https://doi.org/10.1016/j.molcel.2015.11.006
PMid:26590712
Huang J., Zhang X., Zhang M., Zhu J.D., Zhang Y.L., Lin Y., Wang K.S., Qi X.F., Zhang Q., Liu G.Z., Yu J., Cui Y., Yang P.Y., Wang Z.Q., and Han Z.G., 2007, Up-regulation of DLK1 as an imprinted gene could contribute to human hepatocellular carcinoma, Carcinogenesis, 28(5): 1094-1103
https://doi.org/10.1093/carcin/bgl215
PMid:17114643
Lai P.B., Chi T.Y., and Chen G.G., 2007, Different levels of p53 induced either apoptosis or cell cycle arrest in a doxycycline-regulated hepatocellular carcinoma cell line in vitro, Apoptosis, 12(2): 387-393
https://doi.org/10.1007/s10495-006-0571-1
PMid:17191126
Liu Z., Ye Q., Wu L., Gao F., Xie H., Zhou L., Zheng S., and Xu X., 2018, Metallothionein 1 family profiling identifies MT1X as a tumor suppressor involved in the progression and metastastatic capacity of hepatocellular carcinoma, Molecular Carcinogenesis, 57(11): 1435-1444
https://doi.org/10.1002/mc.22846
PMid:29873415
Luengo A., Gui D.Y., and Vander Heiden M.G., 2017, Targeting metabolism for cancer therapy, Cell Chemical Biology, 24(9): 1161-1180
https://doi.org/10.1016/j.chembiol.2017.08.028
PMid:28938091 PMCid:PMC5744685
Mihlan M., Hebecker M., Dahse H.M., Hälbich S., Huber-Lang M., Dahse R., Zipfel P.F., and Józsi M., 2009, Human complement factor H-related protein 4 binds and recruits native pentameric C-reactive protein to necrotic cells, Molecular Immunology, 46(3): 335-344
https://doi.org/10.1016/j.molimm.2008.10.029
PMid:19084272
Monte M.J., Fernandez-Tagarro M., Macias R.I., Jimenez F., Gonzalez-San Martin F., and Marin J.J., 2005, Changes in the expression of genes related to bile acid synthesis and transport by the rat liver during hepatocarcinogenesis, Clinical Science (London, England: 1979), 109(2): 199-207
https://doi.org/10.1042/CS20050035
PMid:15853769
Reig M., Mariño Z., Perelló C., Iñarrairaegui M., Ribeiro A., Lens S., Díaz A., Vilana R.,Darnell A., Varela M., Sangro B., Calleja J.L., Forns X., and Bruix J., 2016, Unexpected high rate of early tumor recurrence in patients with HCV-related HCC undergoing interferon-free therapy, Journal of Hepatology, 65(4): 719-726
https://doi.org/10.1016/j.jhep.2016.04.008
PMid:27084592
Ren J., Chen G.G., Liu Y., Su X., Hu B., Leung B.C., Wang Y., Ho R.L., Yang S., Lu G., Lee C.G., and Lai P.B., 2016, Cytochrome P450 1A2 Metabolizes 17β-Estradiol to Suppress Hepatocellular Carcinoma, PLoS One, 11(4): e0153863
https://doi.org/10.1371/journal.pone.0153863
PMid:27093553 PMCid:PMC4836701
Rossetto A., De Re V., Steffan A., Ravaioli M., Miolo G., Leone P., Racanelli V., Uzzau A., Baccarani U., and Cescon M., 2019, Carcinogenesis and Metastasis in Liver: Cell Physiological Basis, Cancers (Basel). 2019, 11(11): e1731
https://doi.org/10.3390/cancers11111731
PMid:31694274 PMCid:PMC6895858
Seol H.S., Lee S.E., Song J.S., Rhee J.K., Singh S.R., Chang S., and Jang S.J., 2016, Complement proteins C7 and CFH control the stemness of liver cancer cells via LSF-1, Cancer Letters, 372(1): 24-35
https://doi.org/10.1016/j.canlet.2015.12.005
PMid:26723877 PMCid:PMC4744505
Shen J., Yu S., Sun X., Yin M., Fei J., and Zhou J., 2019, Identification of key biomarkers associated with development and prognosis in patients with ovarian carcinoma: evidence from bioinformatic analysis, Journal of Ovarian Research, 12(1): 110
https://doi.org/10.1186/s13048-019-0578-1
PMid:31729978 PMCid:PMC6857166
Suner A., Carr B.I., Akkiz H., Uskudar O., Kuran S., Tokat Y., Tokmak S., Ballı T., Ulku A., AkCam T., Delik A., Arslan B., Doran F., YalCın K., Ekinci N., Yilmaz S., Ozakyol A., Yücesoy M., BahCeci H.I., Polat K.Y., Şimsek H., Ormeci N., Sonsuz A., Demir M., KılıC M., Uygun A., Demir A., Altıntas E., Karakulah G., Temel T., and Bektas A., 2018, Inflammatory markers C-reactive protein and PLR in relation to HCC characteristics, Journal of Translational Science, 5(3): 1-6
https://doi.org/10.15761/JTS.1000260
PMid:30662766 PMCid:PMC6333412
Tang Z., Li C., Kang B., Gao G., Li C., and Zhang Z., 2017, GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses, Nucleic Acids Research, 45(W1): W98-W102
https://doi.org/10.1093/nar/gkx247
PMid:28407145 PMCid:PMC5570223
Wong N., Yeo W., Wong W.L., Wong N.L., Chan K.Y., Mo F.K., Koh J., Chan S.L., Chan A.T., Lai P.B., Ching A.K., Tong J.H., Ng H.K., Johnson P.J., and To K.F., 2009, TOP2A overexpression in hepatocellular carcinoma correlates with early age onset, shorter patients survival and chemoresistance, International Journal of Cancer, 124(3): 644-652
https://doi.org/10.1002/ijc.23968
PMid:19003983
Wu C.X., Wang X.Q., Chok S.H., Man K., Tsang S.H.Y., Chan A.C.Y., Ma K.W., Xia W., and Cheung T.T., 2018, Blocking CDK1/PDK1/β-Catenin signaling by CDK1 inhibitor RO3306 increased the efficacy of sorafenib treatment by targeting cancer stem cells in a preclinical model of hepatocellular carcinoma, Theranostics, 8(14): 3737-3750
https://doi.org/10.7150/thno.25487
PMid:30083256 PMCid:PMC6071527
Wu J.M., Skill N.J., and Maluccio M.A., 2010, Evidence of aberrant lipid metabolism in hepatitis C and hepatocellular carcinoma, HPB (Oxford), 12(9): 625-636
https://doi.org/10.1111/j.1477-2574.2010.00207.x
PMid:20961371 PMCid:PMC2999790
Xia H., Kong S.N., Chen J., Shi M., Sekar K., Seshachalam V.P., Rajasekaran M., Goh B.K.P., Ooi L.L., and Hui K.M., 2016, MELK is an oncogenic kinase essential for early hepatocellular carcinoma recurrence, Cancer Letters, 383(1): 85-93
https://doi.org/10.1016/j.canlet.2016.09.017
PMid:27693640
Ye Z., Wang F., Yan F., Wang L., Li B., Liu T., Hu F., Jiang M., Li W., and Fu Z., 2019, Bioinformatic identification of candidate biomarkers and related transcription factors in nasopharyngeal carcinoma, World Journal of Surgical Oncology, 17(1): 60
https://doi.org/10.1186/s12957-019-1605-9
PMid:30935420 PMCid:PMC6444505
Yu D., Green B., Marrone A., Guo Y., Kadlubar S., Lin D., Fuscoe J., Pogribny I., and Ning B., 2015, Suppression of CYP2C9 by microRNA hsa-miR-128-3p in human liver cells and association with hepatocellular carcinoma, Scientific Reports, 5: 8534
https://doi.org/10.1038/srep08534
PMid:25704921 PMCid:PMC4336941
Yu H.Y., Liu S.M., and Li B., 2019, Diagnostic performance of combined detection of AFP, AFU and TSGF in early liver cancer, Aizheng Jinzhan (Oncology Progress), 17(15): 1789-1791+1842
Yuan T., Chen Z., Yan F., Qian M., Luo H., Ye S., Cao J., Ying M., Dai X., Gai R., Yang B., He Q., and Zhu H., 2019, Deubiquitinating enzyme USP10 promotes hepatocellular carcinoma metastasis through deubiquitinating and stabilizing Smad4 protein, Molecular Oncology, 14(1): 197-210
https://doi.org/10.1002/1878-0261.12596
PMid:31721429 PMCid:PMC6944132
Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., and Chanda S.K., 2019, Metascape provides a biologist-oriented resource for the analysis of systems-level datasets, Nature Communications, 10(1): 1523
https://doi.org/10.1038/s41467-019-09234-6
PMid:30944313 PMCid:PMC6447622
Associated material
. Readers' comments
Other articles by authors
. Lin Qiu
. Jian Huang
Related articles
. Hepatocellular carcinoma
. Differentially expressed genes
. Biomarkers
Tools
. Post a comment
.png)
.png)
.png)
.png)